Note
Click here to download the full example code or to run this example in your browser via Binder
Basic nilearn example: manipulating and looking at data#
A simple example showing how to load an existing Nifti file and use basic nilearn functionalities.
# Let us use a Nifti file that is shipped with nilearn
from nilearn.datasets import MNI152_FILE_PATH
# Note that the variable MNI152_FILE_PATH is just a path to a Nifti file
print('Path to MNI152 template: %r' % MNI152_FILE_PATH)
Path to MNI152 template: '/home/alexis/miniconda3/envs/nilearn/lib/python3.10/site-packages/nilearn/datasets/data/mni_icbm152_t1_tal_nlin_sym_09a_converted.nii.gz'
A first step: looking at our data#
Let’s quickly plot this file:
from nilearn import plotting
plotting.plot_img(MNI152_FILE_PATH)
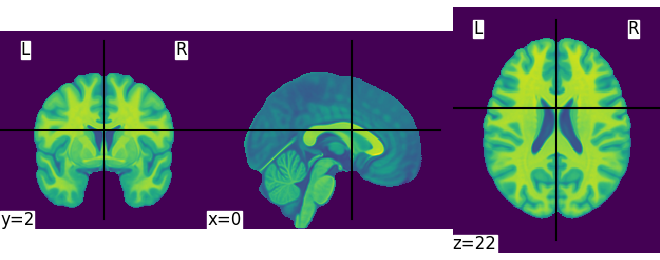
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7ff8fb1df640>
This is not a very pretty plot. We just used the simplest possible code. There is a whole section of the documentation on making prettier code.
Exercise: Try plotting one of your own files. In the above, MNI152_FILE_PATH is nothing more than a string with a path pointing to a nifti image. You can replace it with a string pointing to a file on your disk. Note that it should be a 3D volume, and not a 4D volume.
Simple image manipulation: smoothing#
Let’s use an image-smoothing function from nilearn:
nilearn.image.smooth_img
Functions containing ‘img’ can take either a filename or an image as input.
Here we give as inputs the image filename and the smoothing value in mm
from nilearn import image
smooth_anat_img = image.smooth_img(MNI152_FILE_PATH, fwhm=3)
# While we are giving a file name as input, the function returns
# an in-memory object:
smooth_anat_img
<nibabel.nifti1.Nifti1Image object at 0x7ff8fd58ec50>
This is an in-memory object. We can pass it to nilearn function, for instance to look at it
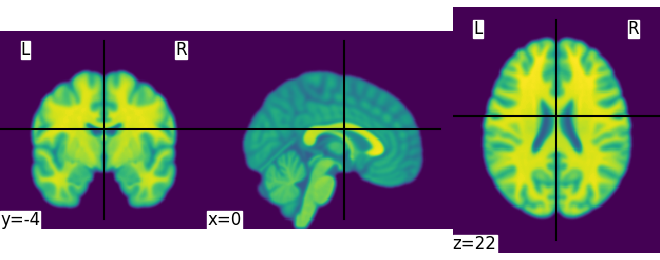
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7ff8fb1dfe50>
We could also pass it to the smoothing function
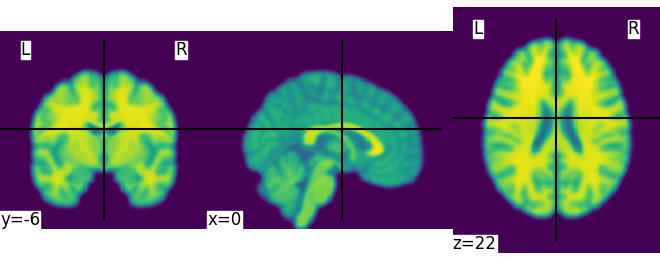
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7ff8fad39d50>
Saving results to a file#
We can save any in-memory object as follows:
more_smooth_anat_img.to_filename('more_smooth_anat_img.nii.gz')
Finally, calling plotting.show() is necessary to display the figure when running as a script outside IPython
To recap, all the nilearn tools can take data as filenames or in-memory objects, and return brain volumes as in-memory objects. These can be passed on to other nilearn tools, or saved to disk.
Total running time of the script: ( 0 minutes 5.321 seconds)
Estimated memory usage: 170 MB