Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.datasets.fetch_atlas_yeo_2011#
- nilearn.datasets.fetch_atlas_yeo_2011(data_dir=None, url=None, resume=True, verbose=1)[source]#
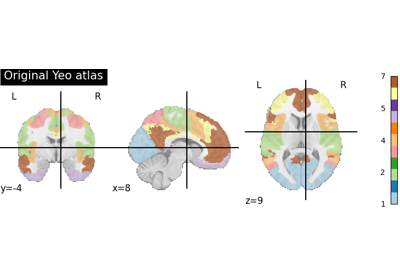
Download and return file names for the Yeo 2011 parcellation.
This function retrieves the so-called yeo deterministic atlases. The provided images are in MNI152 space and have shapes equal to
(256, 256, 256, 1). They contain consecutive integers values from 0 (background) to either 7 or 17 depending on the atlas version considered.For more information on this dataset’s structure, see 1, and 2.
- Parameters
- data_dir
pathlib.Pathorstr, optional Path where data should be downloaded. By default, files are downloaded in home directory.
- url
str, optional URL of file to download. Override download URL. Used for test only (or if you setup a mirror of the data). Default=None.
- resume
bool, optional Whether to resume download of a partly-downloaded file. Default=True.
- verbose
int, optional Verbosity level (0 means no message). Default=1.
- data_dir
- Returns
- data
sklearn.utils.Bunch Dictionary-like object, keys are:
‘thin_7’:
str, path to nifti file containing the 7 regions parcellation fitted to thin template cortex segmentations. The image contains integer values which can be interpreted as the indices incolors_7.‘thick_7’:
str, path to nifti file containing the 7 region parcellation fitted to thick template cortex segmentations. The image contains integer values which can be interpreted as the indices incolors_7.‘thin_17’:
str, path to nifti file containing the 17 region parcellation fitted to thin template cortex segmentations. The image contains integer values which can be interpreted as the indices incolors_17.‘thick_17’:
str, path to nifti file containing the 17 region parcellation fitted to thick template cortex segmentations. The image contains integer values which can be interpreted as the indices incolors_17.‘colors_7’:
str, path to colormaps text file for 7 region parcellation. This file maps voxel integer values fromdata.thin_7anddata.tick_7to network names.‘colors_17’:
str, path to colormaps text file for 17 region parcellation. This file maps voxel integer values fromdata.thin_17anddata.tick_17to network names.‘anat’:
str, path to nifti file containing the anatomy image.‘description’:
str, description of the atlas.
- data
Notes
Licence: unknown.
References
- 1
Cortical parcellation estimated by intrinsic functional connectivity. http://surfer.nmr.mgh.harvard.edu/fswiki/CorticalParcellation_Yeo2011. Accessed: 2021-05-19.
- 2
B. T. Thomas Yeo, Fenna M. Krienen, Jorge Sepulcre, Mert R. Sabuncu, Danial Lashkari, Marisa Hollinshead, Joshua L. Roffman, Jordan W. Smoller, Lilla Zöllei, Jonathan R. Polimeni, Bruce Fischl, Hesheng Liu, and Randy L. Buckner. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of Neurophysiology, 106(3):1125–1165, 2011. PMID: 21653723. URL: https://doi.org/10.1152/jn.00338.2011, arXiv:https://doi.org/10.1152/jn.00338.2011, doi:10.1152/jn.00338.2011.
Examples using nilearn.datasets.fetch_atlas_yeo_2011#
Comparing connectomes on different reference atlases