Note
Click here to download the full example code or to run this example in your browser via Binder
Example of surface-based first-level analysis#
A full step-by-step example of fitting a GLM to experimental data sampled on the cortical surface and visualizing the results.
More specifically:
A sequence of fMRI volumes is loaded.
fMRI data are projected onto a reference cortical surface (the FreeSurfer template, fsaverage).
A design matrix describing all the effects related to the data is computed.
A GLM is applied to the dataset (effect/covariance, then contrast estimation).
The result of the analysis are statistical maps that are defined on the brain mesh. We display them using Nilearn capabilities.
The projection of fMRI data onto a given brain mesh requires that both are initially defined in the same space.
The functional data should be coregistered to the anatomy from which the mesh was obtained.
Another possibility, used here, is to project the normalized fMRI data to an MNI-coregistered mesh, such as fsaverage.
The advantage of this second approach is that it makes it easy to run second-level analyses on the surface. On the other hand, it is obviously less accurate than using a subject-tailored mesh.
Prepare data and analysis parameters#
Prepare the timing parameters.
t_r = 2.4
slice_time_ref = 0.5
Prepare the data. First, the volume-based fMRI data.
from nilearn.datasets import fetch_localizer_first_level
data = fetch_localizer_first_level()
fmri_img = data.epi_img
Second, the experimental paradigm.
events_file = data.events
import pandas as pd
events = pd.read_table(events_file)
Project the fMRI image to the surface#
For this we need to get a mesh representing the geometry of the surface. We could use an individual mesh, but we first resort to a standard mesh, the so-called fsaverage5 template from the FreeSurfer software.
import nilearn
fsaverage = nilearn.datasets.fetch_surf_fsaverage()
The projection function simply takes the fMRI data and the mesh. Note that those correspond spatially, are they are both in MNI space.
from nilearn import surface
texture = surface.vol_to_surf(fmri_img, fsaverage.pial_right)
Perform first level analysis#
This involves computing the design matrix and fitting the model. We start by specifying the timing of fMRI frames.
import numpy as np
n_scans = texture.shape[1]
frame_times = t_r * (np.arange(n_scans) + .5)
Create the design matrix.
We specify an hrf model containing the Glover model and its time derivative The drift model is implicitly a cosine basis with a period cutoff at 128s.
from nilearn.glm.first_level import make_first_level_design_matrix
design_matrix = make_first_level_design_matrix(frame_times,
events=events,
hrf_model='glover + derivative'
)
Setup and fit GLM.
Note that the output consists in 2 variables: labels and fit. labels tags voxels according to noise autocorrelation. estimates contains the parameter estimates. We keep them for later contrast computation.
Estimate contrasts#
Specify the contrasts.
For practical purpose, we first generate an identity matrix whose size is the number of columns of the design matrix.
At first, we create basic contrasts.
basic_contrasts = dict([(column, contrast_matrix[i])
for i, column in enumerate(design_matrix.columns)])
Next, we add some intermediate contrasts and one contrast adding all conditions with some auditory parts.
basic_contrasts['audio'] = (
basic_contrasts['audio_left_hand_button_press']
+ basic_contrasts['audio_right_hand_button_press']
+ basic_contrasts['audio_computation']
+ basic_contrasts['sentence_listening'])
# one contrast adding all conditions involving instructions reading
basic_contrasts['visual'] = (
basic_contrasts['visual_left_hand_button_press']
+ basic_contrasts['visual_right_hand_button_press']
+ basic_contrasts['visual_computation']
+ basic_contrasts['sentence_reading'])
# one contrast adding all conditions involving computation
basic_contrasts['computation'] = (basic_contrasts['visual_computation']
+ basic_contrasts['audio_computation'])
# one contrast adding all conditions involving sentences
basic_contrasts['sentences'] = (basic_contrasts['sentence_listening']
+ basic_contrasts['sentence_reading'])
Finally, we create a dictionary of more relevant contrasts
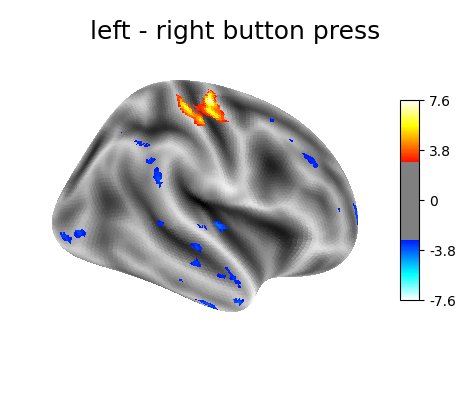
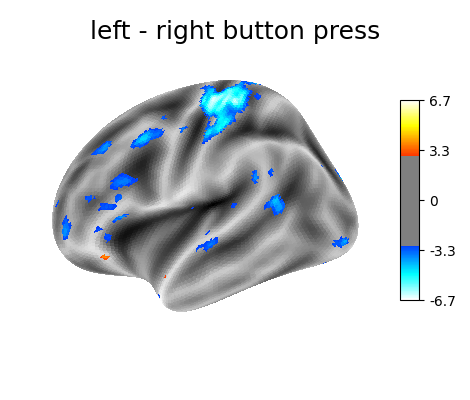
‘left - right button press’: probes motor activity in left versus right button presses.
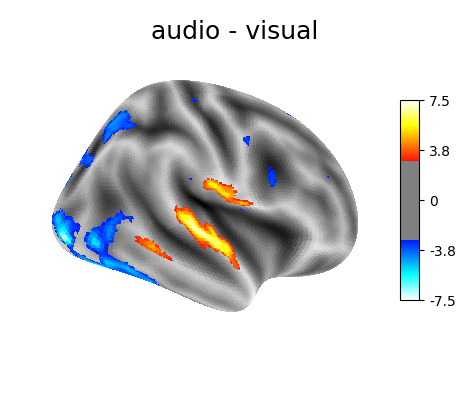
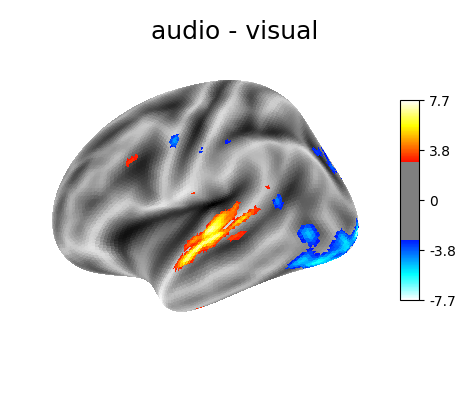
‘audio - visual’: probes the difference of activity between listening to some content or reading the same type of content (instructions, stories).
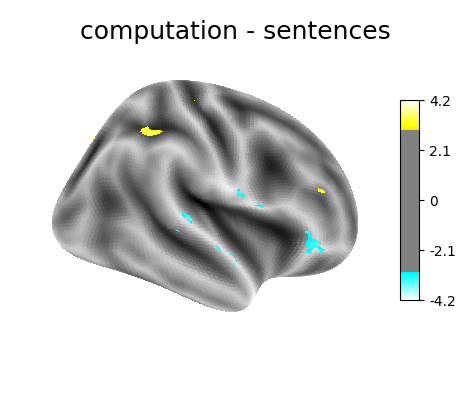
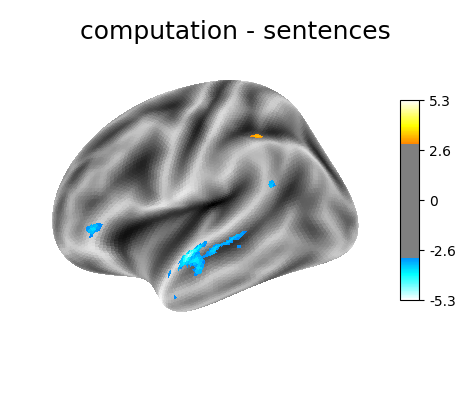
‘computation - sentences’: looks at the activity when performing a mental computation task versus simply reading sentences.
Of course, we could define other contrasts, but we keep only 3 for simplicity.
contrasts = {
'left - right button press': (
basic_contrasts['audio_left_hand_button_press']
- basic_contrasts['audio_right_hand_button_press']
+ basic_contrasts['visual_left_hand_button_press']
- basic_contrasts['visual_right_hand_button_press']
),
'audio - visual': basic_contrasts['audio'] - basic_contrasts['visual'],
'computation - sentences': (basic_contrasts['computation'] -
basic_contrasts['sentences']
)
}
Let’s estimate the contrasts by iterating over them.
from nilearn.glm.contrasts import compute_contrast
from nilearn import plotting
for index, (contrast_id, contrast_val) in enumerate(contrasts.items()):
print(' Contrast % i out of %i: %s, right hemisphere' %
(index + 1, len(contrasts), contrast_id))
# compute contrast-related statistics
contrast = compute_contrast(labels, estimates, contrast_val,
contrast_type='t')
# we present the Z-transform of the t map
z_score = contrast.z_score()
# we plot it on the surface, on the inflated fsaverage mesh,
# together with a suitable background to give an impression
# of the cortex folding.
plotting.plot_surf_stat_map(
fsaverage.infl_right, z_score, hemi='right',
title=contrast_id, colorbar=True,
threshold=3., bg_map=fsaverage.sulc_right)
Contrast 1 out of 3: left - right button press, right hemisphere
Contrast 2 out of 3: audio - visual, right hemisphere
Contrast 3 out of 3: computation - sentences, right hemisphere
Analysing the left hemisphere#
Note that re-creating the above analysis for the left hemisphere requires little additional code!
We project the fMRI data to the mesh.
Then we estimate the General Linear Model.
Finally, we create contrast-specific maps and plot them.
for index, (contrast_id, contrast_val) in enumerate(contrasts.items()):
print(' Contrast % i out of %i: %s, left hemisphere' %
(index + 1, len(contrasts), contrast_id))
# compute contrasts
contrast = compute_contrast(labels, estimates, contrast_val,
contrast_type='t')
z_score = contrast.z_score()
# plot the result
plotting.plot_surf_stat_map(
fsaverage.infl_left, z_score, hemi='left',
title=contrast_id, colorbar=True,
threshold=3., bg_map=fsaverage.sulc_left)
plotting.show()
Contrast 1 out of 3: left - right button press, left hemisphere
Contrast 2 out of 3: audio - visual, left hemisphere
Contrast 3 out of 3: computation - sentences, left hemisphere
Total running time of the script: ( 0 minutes 15.307 seconds)
Estimated memory usage: 209 MB