Note
Click here to download the full example code or to run this example in your browser via Binder
Advanced decoding using scikit learn#
This tutorial opens the box of decoding pipelines to bridge integrated
functionalities provided by the nilearn.decoding.Decoder object
with more advanced usecases. It reproduces basic examples functionalities with
direct calls to scikit-learn function and gives pointers to more advanced
objects. If some concepts seem unclear, please refer to the documentation on decoding
and in particular to the advanced section.
As in many other examples, we perform decoding of the visual category of a
stimuli on Haxby 2001 dataset, focusing on distinguishing two categories :
face and cat images.
J.V. Haxby et al. “Distributed and Overlapping Representations of Faces and Objects in Ventral Temporal Cortex”, Science vol 293 (2001), p 2425.-2430.
Note
If you are using Nilearn with a version older than 0.9.0,
then you should either upgrade your version or import maskers
from the input_data module instead of the maskers module.
That is, you should manually replace in the following example all occurrences of:
from nilearn.maskers import NiftiMasker
with:
from nilearn.input_data import NiftiMasker
Retrieve and load the fMRI data from the Haxby study#
First download the data#
# The :func:`nilearn.datasets.fetch_haxby` function will download the
# Haxby dataset composed of fmri images in a Niimg, a spatial mask and a text
# document with label of each image
from nilearn import datasets
haxby_dataset = datasets.fetch_haxby()
mask_filename = haxby_dataset.mask_vt[0]
fmri_filename = haxby_dataset.func[0]
# Loading the behavioral labels
import pandas as pd
behavioral = pd.read_csv(haxby_dataset.session_target[0], delimiter=' ')
behavioral
We keep only a images from a pair of conditions(cats versus faces).
from nilearn.image import index_img
conditions = behavioral['labels']
condition_mask = conditions.isin(['face', 'cat'])
fmri_niimgs = index_img(fmri_filename, condition_mask)
conditions = conditions[condition_mask]
# Convert to numpy array
conditions = conditions.values
session_label = behavioral['chunks'][condition_mask]
Performing decoding with scikit-learn#
# Importing a classifier
# ........................
# We can import many predictive models from scikit-learn that can be used in a
# decoding pipelines. They are all used with the same `fit()` and `predict()`
# functions. Let's define a `Support Vector Classifier <http://scikit-learn.org/stable/modules/svm.html >`_ (or SVC).
from sklearn.svm import SVC
svc = SVC()
Masking the data#
To use a scikit-learn estimator on brain images, you should first mask the
data using a nilearn.maskers.NiftiMasker to extract only the
voxels inside the mask of interest, and transform 4D input fMRI data to
2D arrays(shape=(n_timepoints, n_voxels)) that estimators can work on.
from nilearn.maskers import NiftiMasker
masker = NiftiMasker(mask_img=mask_filename, runs=session_label,
smoothing_fwhm=4, standardize=True,
memory="nilearn_cache", memory_level=1)
fmri_masked = masker.fit_transform(fmri_niimgs)
/home/alexis/miniconda3/envs/nilearn/lib/python3.10/site-packages/nilearn/image/resampling.py:452: UserWarning: The provided image has no sform in its header. Please check the provided file. Results may not be as expected.
warnings.warn("The provided image has no sform in its header. "
Cross-validation with scikit-learn#
To train and test the model in a meaningful way we use cross-validation with
the function sklearn.model_selection.cross_val_score that computes
for you the score for the different folds of cross-validation.
from sklearn.model_selection import cross_val_score
# Here `cv=5` stipulates a 5-fold cross-validation
cv_scores = cross_val_score(svc, fmri_masked, conditions, cv=5)
print("SVC accuracy: {:.3f}".format(cv_scores.mean()))
SVC accuracy: 0.823
Tuning cross-validation parameters#
You can change many parameters of the cross_validation here, for example:
use a different cross - validation scheme, for example LeaveOneGroupOut()
speed up the computation by using n_jobs = -1, which will spread the computation equally across all processors.
use a different scoring function, as a keyword or imported from scikit-learn : scoring = ‘roc_auc’
from sklearn.model_selection import LeaveOneGroupOut
cv = LeaveOneGroupOut()
cv_scores = cross_val_score(svc, fmri_masked, conditions, cv=cv,
scoring='roc_auc', groups=session_label, n_jobs=-1)
print("SVC accuracy (tuned parameters): {:.3f}".format(cv_scores.mean()))
SVC accuracy (tuned parameters): 0.858
Measuring the chance level#
sklearn.dummy.DummyClassifier (purely random) estimators are the
simplest way to measure prediction performance at chance. A more controlled
way, but slower, is to do permutation testing on the labels, with
sklearn.model_selection.permutation_test_score.
Dummy estimator#
from sklearn.dummy import DummyClassifier
null_cv_scores = cross_val_score(
DummyClassifier(), fmri_masked, conditions, cv=cv, groups=session_label)
print("Dummy accuracy: {:.3f}".format(null_cv_scores.mean()))
Dummy accuracy: 0.500
Permutation test#
from sklearn.model_selection import permutation_test_score
null_cv_scores = permutation_test_score(
svc, fmri_masked, conditions, cv=cv, groups=session_label)[1]
print("Permutation test score: {:.3f}".format(null_cv_scores.mean()))
Permutation test score: 0.502
Decoding without a mask: Anova-SVM in scikit-lean#
We can also implement feature selection before decoding as a scikit-learn
pipeline`(:class:`sklearn.pipeline.Pipeline). For this, we need to import
the sklearn.feature_selection module and use
sklearn.feature_selection.f_classif, a simple F-score
based feature selection (a.k.a. Anova),
from sklearn.pipeline import Pipeline
from sklearn.feature_selection import SelectPercentile, f_classif
from sklearn.model_selection import cross_validate
from sklearn.svm import LinearSVC
feature_selection = SelectPercentile(f_classif, percentile=10)
anova_svc = Pipeline([('anova', feature_selection), ('svc', LinearSVC())])
# We can use our ``anova_svc`` object exactly as we were using our ``svc``
# object previously.
# As we want to investigate our model, we use sklearn `cross_validate` function
# with `return_estimator = True` instead of cross_val_score, to save the estimator
fitted_pipeline = cross_validate(anova_svc, fmri_masked, conditions,
cv=cv, groups=session_label, return_estimator=True)
print(
"ANOVA+SVC test score: {:.3f}".format(fitted_pipeline["test_score"].mean()))
ANOVA+SVC test score: 0.801
Visualize the ANOVA + SVC’s discriminating weights#
# retrieve the pipeline fitted on the first cross-validation fold and its SVC
# coefficients
first_pipeline = fitted_pipeline["estimator"][0]
svc_coef = first_pipeline.named_steps['svc'].coef_
print("After feature selection, the SVC is trained only on {} features".format(
svc_coef.shape[1]))
# We invert the feature selection step to put these coefs in the right 2D place
full_coef = first_pipeline.named_steps['anova'].inverse_transform(svc_coef)
print("After inverting feature selection, we have {} features back".format(
full_coef.shape[1]))
# We apply the inverse of masking on these to make a 4D image that we can plot
from nilearn.plotting import plot_stat_map
weight_img = masker.inverse_transform(full_coef)
plot_stat_map(weight_img, title='Anova+SVC weights')
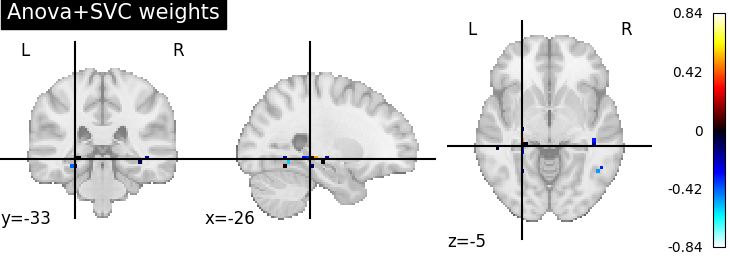
After feature selection, the SVC is trained only on 47 features
After inverting feature selection, we have 464 features back
/home/alexis/miniconda3/envs/nilearn/lib/python3.10/site-packages/nilearn/plotting/img_plotting.py:300: FutureWarning: Default resolution of the MNI template will change from 2mm to 1mm in version 0.10.0
anat_img = load_mni152_template()
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f55f2b161d0>
Going further with scikit-learn#
Changing the prediction engine#
To change the prediction engine, we just need to import it and use in our pipeline instead of the SVC. We can try Fisher’s Linear Discriminant Analysis (LDA)
# Construct the new estimator object and use it in a new pipeline after anova
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
feature_selection = SelectPercentile(f_classif, percentile=10)
lda = LinearDiscriminantAnalysis()
anova_lda = Pipeline([('anova', feature_selection), ('LDA', lda)])
# Recompute the cross-validation score:
import numpy as np
cv_scores = cross_val_score(anova_lda, fmri_masked,
conditions, cv=cv, verbose=1, groups=session_label)
classification_accuracy = np.mean(cv_scores)
n_conditions = len(set(conditions)) # number of target classes
print("ANOVA + LDA classification accuracy: %.4f / Chance Level: %.4f" %
(classification_accuracy, 1. / n_conditions))
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.1s finished
ANOVA + LDA classification accuracy: 0.8009 / Chance Level: 0.5000
Changing the feature selection#
Let’s say that you want a more sophisticated feature selection, for example a Recursive Feature Elimination(RFE) before a svc. We follows the same principle.
# Import it and define your fancy objects
from sklearn.feature_selection import RFE
svc = SVC()
rfe = RFE(SVC(kernel='linear', C=1.), n_features_to_select=50, step=0.25)
# Create a new pipeline, composing the two classifiers `rfe` and `svc`
rfe_svc = Pipeline([('rfe', rfe), ('svc', svc)])
# Recompute the cross-validation score
# cv_scores = cross_val_score(rfe_svc, fmri_masked, target, cv=cv, n_jobs=-1, verbose=1)
# But, be aware that this can take * A WHILE * ...
Total running time of the script: ( 0 minutes 11.315 seconds)
Estimated memory usage: 928 MB