Note
Click here to download the full example code or to run this example in your browser via Binder
Example of pattern recognition on simulated data#
This example simulates data according to a very simple sketch of brain imaging data and applies machine learning techniques to predict output values.
We use a very simple generating function to simulate data, as in Michel et al. 2012 , a linear model with a random design matrix X:
\mathbf{y} = \mathbf{X} \mathbf{w} + \mathbf{e}
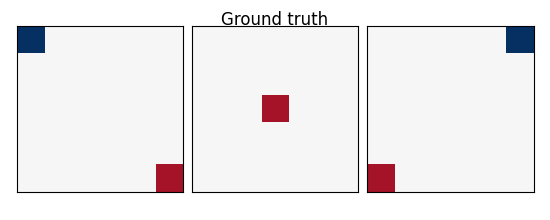
w: the weights of the linear model correspond to the predictive brain regions. Here, in the simulations, they form a 3D image with 5, four of which in opposite corners and one in the middle, as plotted below.
X: the design matrix corresponds to the observed fMRI data. Here we simulate random normal variables and smooth them as in Gaussian fields.
e is random normal noise.
# Licence : BSD
print(__doc__)
from time import time
import numpy as np
import matplotlib.pyplot as plt
from scipy import linalg
from scipy.ndimage import gaussian_filter
from sklearn import linear_model, svm
from sklearn.utils import check_random_state
from sklearn.model_selection import KFold
from sklearn.feature_selection import f_regression
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import make_pipeline
import nibabel
from nilearn import decoding
import nilearn.masking
from nilearn.plotting import show
A function to generate data#
def create_simulation_data(snr=0, n_samples=2 * 100, size=12, random_state=1):
generator = check_random_state(random_state)
roi_size = 2 # size / 3
smooth_X = 1
# Coefs
w = np.zeros((size, size, size))
w[0:roi_size, 0:roi_size, 0:roi_size] = -0.6
w[-roi_size:, -roi_size:, 0:roi_size] = 0.5
w[0:roi_size, -roi_size:, -roi_size:] = -0.6
w[-roi_size:, 0:roi_size:, -roi_size:] = 0.5
w[(size - roi_size) // 2:(size + roi_size) // 2,
(size - roi_size) // 2:(size + roi_size) // 2,
(size - roi_size) // 2:(size + roi_size) // 2] = 0.5
w = w.ravel()
# Generate smooth background noise
XX = generator.randn(n_samples, size, size, size)
noise = []
for i in range(n_samples):
Xi = gaussian_filter(XX[i, :, :, :], smooth_X)
Xi = Xi.ravel()
noise.append(Xi)
noise = np.array(noise)
# Generate the signal y
y = generator.randn(n_samples)
X = np.dot(y[:, np.newaxis], w[np.newaxis])
norm_noise = linalg.norm(X, 2) / np.exp(snr / 20.)
noise_coef = norm_noise / linalg.norm(noise, 2)
noise *= noise_coef
snr = 20 * np.log(linalg.norm(X, 2) / linalg.norm(noise, 2))
print("SNR: %.1f dB" % snr)
# Mixing of signal + noise and splitting into train/test
X += noise
X -= X.mean(axis=-1)[:, np.newaxis]
X /= X.std(axis=-1)[:, np.newaxis]
X_test = X[n_samples // 2:, :]
X_train = X[:n_samples // 2, :]
y_test = y[n_samples // 2:]
y = y[:n_samples // 2]
return X_train, X_test, y, y_test, snr, w, size
A simple function to plot slices#
def plot_slices(data, title=None):
plt.figure(figsize=(5.5, 2.2))
vmax = np.abs(data).max()
for i in (0, 6, 11):
plt.subplot(1, 3, i // 5 + 1)
plt.imshow(data[:, :, i], vmin=-vmax, vmax=vmax,
interpolation="nearest", cmap=plt.cm.RdBu_r)
plt.xticks(())
plt.yticks(())
plt.subplots_adjust(hspace=0.05, wspace=0.05, left=.03, right=.97, top=.9)
if title is not None:
plt.suptitle(title, y=.95)
Create data#
X_train, X_test, y_train, y_test, snr, coefs, size = \
create_simulation_data(snr=-10, n_samples=100, size=12)
# Create masks for SearchLight. process_mask is the voxels where SearchLight
# computation is performed. It is a subset of the brain mask, just to reduce
# computation time.
mask = np.ones((size, size, size), dtype=bool)
mask_img = nibabel.Nifti1Image(mask.astype("uint8"), np.eye(4))
process_mask = np.zeros((size, size, size), dtype=bool)
process_mask[:, :, 0] = True
process_mask[:, :, 6] = True
process_mask[:, :, 11] = True
process_mask_img = nibabel.Nifti1Image(process_mask.astype("uint8"), np.eye(4))
coefs = np.reshape(coefs, [size, size, size])
plot_slices(coefs, title="Ground truth")
SNR: -10.0 dB
Run different estimators#
We can now run different estimators and look at their prediction score, as well as the feature maps that they recover. Namely, we will use
A support vector regression (SVM)
An elastic-net
A Bayesian ridge estimator, i.e. a ridge estimator that sets its parameter according to a metaprior
A ridge estimator that set its parameter by cross-validation
Note that the RidgeCV and the ElasticNetCV have names ending in CV that stands for cross-validation: in the list of possible alpha values that they are given, they choose the best by cross-validation.
bayesian_ridge = make_pipeline(
StandardScaler(), linear_model.BayesianRidge()
)
estimators = [
('bayesian_ridge', bayesian_ridge),
('enet_cv', linear_model.ElasticNetCV(alphas=[5, 1, 0.5, 0.1],
l1_ratio=0.05)),
('ridge_cv', linear_model.RidgeCV(alphas=[100, 10, 1, 0.1], cv=5)),
('svr', svm.SVR(kernel='linear', C=0.001)),
('searchlight', decoding.SearchLight(mask_img,
process_mask_img=process_mask_img,
radius=2.7,
scoring='r2',
estimator=svm.SVR(kernel="linear"),
cv=KFold(n_splits=4),
verbose=1,
n_jobs=1,
)
)
]
Run the estimators
As the estimators expose a fairly consistent API, we can all fit them in a for loop: they all have a fit method for fitting the data, a score method to retrieve the prediction score, and because they are all linear models, a coef_ attribute that stores the coefficients w estimated
for name, estimator in estimators:
t1 = time()
if name != "searchlight":
estimator.fit(X_train, y_train)
else:
X = nilearn.masking.unmask(X_train, mask_img)
estimator.fit(X, y_train)
del X
elapsed_time = time() - t1
if name != 'searchlight':
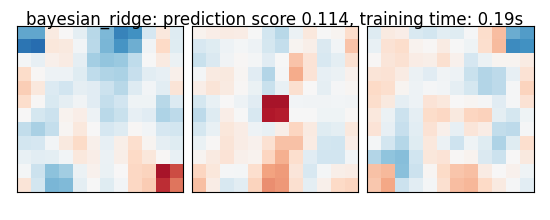
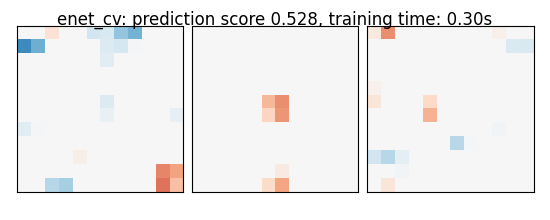
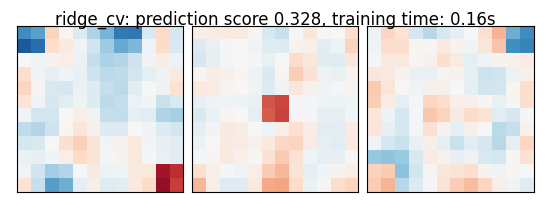
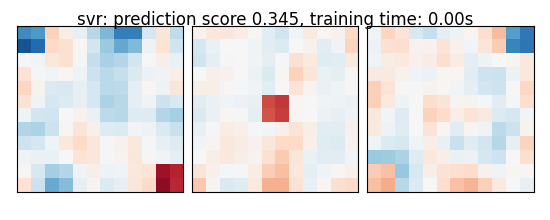
if name == 'bayesian_ridge':
coefs = estimator.named_steps['bayesianridge'].coef_
else:
coefs = estimator.coef_
coefs = np.reshape(coefs, [size, size, size])
score = estimator.score(X_test, y_test)
title = '%s: prediction score %.3f, training time: %.2fs' % (
name, score, elapsed_time)
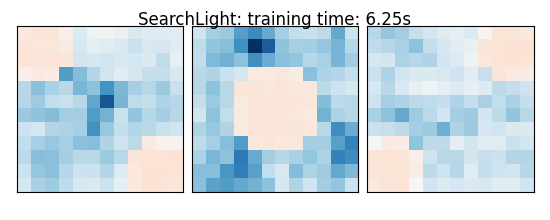
else: # Searchlight
coefs = estimator.scores_
title = '%s: training time: %.2fs' % (
estimator.__class__.__name__,
elapsed_time)
# We use the plot_slices function provided in the example to
# plot the results
plot_slices(coefs, title=title)
print(title)
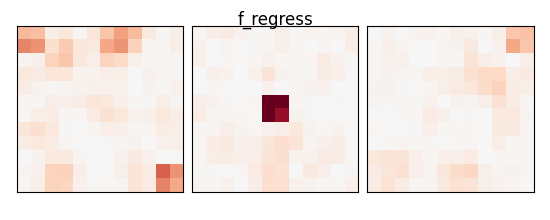
f_values, p_values = f_regression(X_train, y_train)
p_values = np.reshape(p_values, (size, size, size))
p_values = -np.log10(p_values)
p_values[np.isnan(p_values)] = 0
p_values[p_values > 10] = 10
plot_slices(p_values, title="f_regress")
show()
bayesian_ridge: prediction score 0.114, training time: 0.19s
enet_cv: prediction score 0.528, training time: 0.30s
ridge_cv: prediction score 0.328, training time: 0.16s
svr: prediction score 0.345, training time: 0.00s
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
Job #1, processed 0/432 voxels (0.00%, 139 seconds remaining)
Job #1, processed 10/432 voxels (2.31%, 6 seconds remaining)
Job #1, processed 20/432 voxels (4.63%, 5 seconds remaining)
Job #1, processed 30/432 voxels (6.94%, 5 seconds remaining)
Job #1, processed 40/432 voxels (9.26%, 5 seconds remaining)
Job #1, processed 50/432 voxels (11.57%, 5 seconds remaining)
Job #1, processed 60/432 voxels (13.89%, 5 seconds remaining)
Job #1, processed 70/432 voxels (16.20%, 5 seconds remaining)
Job #1, processed 80/432 voxels (18.52%, 5 seconds remaining)
Job #1, processed 90/432 voxels (20.83%, 4 seconds remaining)
Job #1, processed 100/432 voxels (23.15%, 4 seconds remaining)
Job #1, processed 110/432 voxels (25.46%, 4 seconds remaining)
Job #1, processed 120/432 voxels (27.78%, 4 seconds remaining)
Job #1, processed 130/432 voxels (30.09%, 4 seconds remaining)
Job #1, processed 140/432 voxels (32.41%, 4 seconds remaining)
Job #1, processed 150/432 voxels (34.72%, 3 seconds remaining)
Job #1, processed 160/432 voxels (37.04%, 3 seconds remaining)
Job #1, processed 170/432 voxels (39.35%, 3 seconds remaining)
Job #1, processed 180/432 voxels (41.67%, 3 seconds remaining)
Job #1, processed 190/432 voxels (43.98%, 3 seconds remaining)
Job #1, processed 200/432 voxels (46.30%, 3 seconds remaining)
Job #1, processed 210/432 voxels (48.61%, 2 seconds remaining)
Job #1, processed 220/432 voxels (50.93%, 2 seconds remaining)
Job #1, processed 230/432 voxels (53.24%, 2 seconds remaining)
Job #1, processed 240/432 voxels (55.56%, 2 seconds remaining)
Job #1, processed 250/432 voxels (57.87%, 2 seconds remaining)
Job #1, processed 260/432 voxels (60.19%, 2 seconds remaining)
Job #1, processed 270/432 voxels (62.50%, 2 seconds remaining)
Job #1, processed 280/432 voxels (64.81%, 1 seconds remaining)
Job #1, processed 290/432 voxels (67.13%, 1 seconds remaining)
Job #1, processed 300/432 voxels (69.44%, 1 seconds remaining)
Job #1, processed 310/432 voxels (71.76%, 1 seconds remaining)
Job #1, processed 320/432 voxels (74.07%, 1 seconds remaining)
Job #1, processed 330/432 voxels (76.39%, 1 seconds remaining)
Job #1, processed 340/432 voxels (78.70%, 1 seconds remaining)
Job #1, processed 350/432 voxels (81.02%, 1 seconds remaining)
Job #1, processed 360/432 voxels (83.33%, 0 seconds remaining)
Job #1, processed 370/432 voxels (85.65%, 0 seconds remaining)
Job #1, processed 380/432 voxels (87.96%, 0 seconds remaining)
Job #1, processed 390/432 voxels (90.28%, 0 seconds remaining)
Job #1, processed 400/432 voxels (92.59%, 0 seconds remaining)
Job #1, processed 410/432 voxels (94.91%, 0 seconds remaining)
Job #1, processed 420/432 voxels (97.22%, 0 seconds remaining)
Job #1, processed 430/432 voxels (99.54%, 0 seconds remaining)
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 5.8s finished
SearchLight: training time: 6.25s
An exercice to go further#
As an exercice, you can use recursive feature elimination (RFE) with the SVM
Read the object’s documentation to find out how to use RFE.
Performance tip: increase the step parameter, or it will be very slow.
from sklearn.feature_selection import RFE
Total running time of the script: ( 0 minutes 8.962 seconds)
Estimated memory usage: 10 MB