Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.datasets.fetch_atlas_surf_destrieux#
- nilearn.datasets.fetch_atlas_surf_destrieux(data_dir=None, url=None, resume=True, verbose=1)[source]#
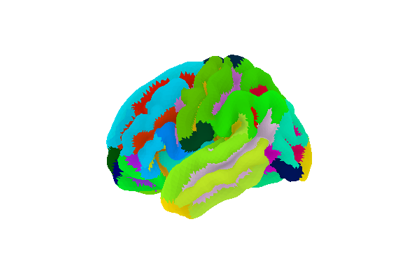
Download and load Destrieux et al, 2010 cortical Deterministic atlas.
See 1.
This atlas returns 76 labels per hemisphere based on sulco-gryal patterns as distributed with Freesurfer in fsaverage5 surface space.
New in version 0.3.
- Parameters
- data_dir
pathlib.Pathorstr, optional Path where data should be downloaded. By default, files are downloaded in home directory.
- url
str, optional URL of file to download. Override download URL. Used for test only (or if you setup a mirror of the data). Default=None.
- resume
bool, optional Whether to resume download of a partly-downloaded file. Default=True.
- verbose
int, optional Verbosity level (0 means no message). Default=1.
- data_dir
- Returns
- data
sklearn.utils.Bunch Dictionary-like object, contains:
‘labels’:
listofstr, list containing the 76 region labels.‘map_left’:
numpy.ndarrayofint, maps each vertex on the left hemisphere of the fsaverage5 surface to its index into the list of label name.‘map_right’:
numpy.ndarrayofint, maps each vertex on the right hemisphere of the fsaverage5 surface to its index into the list of label name.‘description’:
str, description of the dataset.
- data
References
- 1
Christophe Destrieux, Bruce Fischl, Anders Dale, and Eric Halgren. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. NeuroImage, 53(1):1–15, 2010. URL: https://www.sciencedirect.com/science/article/pii/S1053811910008542, doi:https://doi.org/10.1016/j.neuroimage.2010.06.010.