Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.datasets.fetch_atlas_msdl#
- nilearn.datasets.fetch_atlas_msdl(data_dir=None, url=None, resume=True, verbose=1)[source]#
Download and load the MSDL brain Probabilistic atlas.
It can be downloaded at 1, and cited using 2. See also 3 for more information.
- Parameters
- data_dir
pathlib.Pathorstr, optional Path where data should be downloaded. By default, files are downloaded in home directory.
- url
str, optional URL of file to download. Override download URL. Used for test only (or if you setup a mirror of the data). Default=None.
- resume
bool, optional Whether to resume download of a partly-downloaded file. Default=True.
- verbose
int, optional Verbosity level (0 means no message). Default=1.
- data_dir
- Returns
- data
sklearn.utils.Bunch Dictionary-like object, the interest attributes are :
‘maps’:
str, path to nifti file containing the Probabilistic atlas image (shape is equal to(40, 48, 35, 39)).‘labels’:
listofstr, list containing the labels of the regions. There are 39 labels such thatdata.labels[i]corresponds to mapi.‘region_coords’:
listof length-3tuple,data.region_coords[i]contains the coordinates(x, y, z)of regioniin MNI space.‘networks’:
listofstr, list containing the names of the networks. There are 39 network names such thatdata.networks[i]is the network name of regioni.‘description’:
str, description of the atlas.
- data
References
- 1
Spatially constrained parcellation. https://team.inria.fr/parietal/files/2015/01/MSDL_rois.zip. Accessed: 2021-05-19.
- 2
Gael Varoquaux, Alexandre Gramfort, Fabian Pedregosa, Vincent Michel, and Bertrand Thirion. Multi-subject dictionary learning to segment an atlas of brain spontaneous activity. In Information Processing in Medical Imaging, 562–573. Berlin, Heidelberg, 2011. Springer Berlin Heidelberg.
- 3
Gaël Varoquaux and R. Cameron Craddock. Learning and comparing functional connectomes across subjects. NeuroImage, 80:405–415, 2013. Mapping the Connectome. URL: https://www.sciencedirect.com/science/article/pii/S1053811913003340, doi:https://doi.org/10.1016/j.neuroimage.2013.04.007.
Examples using nilearn.datasets.fetch_atlas_msdl#
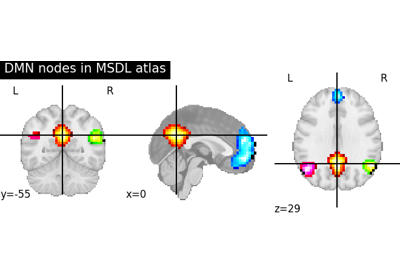
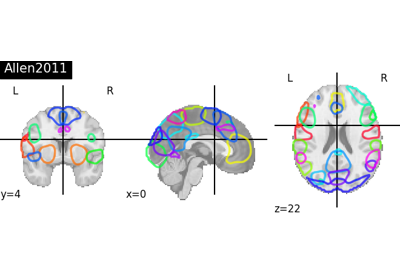
Visualizing a probabilistic atlas: the default mode in the MSDL atlas
Extracting signals of a probabilistic atlas of functional regions
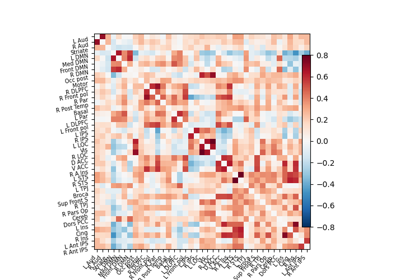
Computing a connectome with sparse inverse covariance
Group Sparse inverse covariance for multi-subject connectome
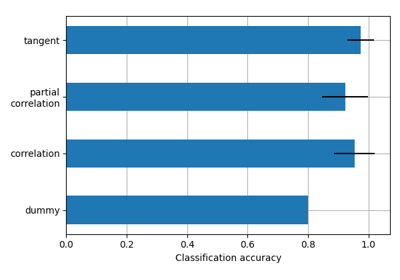
Classification of age groups using functional connectivity