Note
Click here to download the full example code or to run this example in your browser via Binder
Extracting signals of a probabilistic atlas of functional regions#
This example extracts the signal on regions defined via a probabilistic atlas, to construct a functional connectome.
We use the MSDL atlas of functional regions in movie-watching.
The key to extract signals is to use the
nilearn.maskers.NiftiMapsMasker that can transform nifti
objects to time series using a probabilistic atlas.
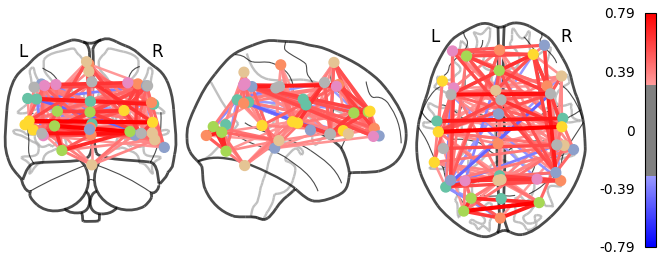
As the MSDL atlas comes with (x, y, z) MNI coordinates for the different regions, we can visualize the matrix as a graph of interaction in a brain. To avoid having too dense a graph, we represent only the 20% edges with the highest values.
Note
If you are using Nilearn with a version older than 0.9.0,
then you should either upgrade your version or import maskers
from the input_data module instead of the maskers module.
That is, you should manually replace in the following example all occurrences of:
from nilearn.maskers import NiftiMasker
with:
from nilearn.input_data import NiftiMasker
Retrieve the atlas and the data#
from nilearn import datasets
atlas = datasets.fetch_atlas_msdl()
# Loading atlas image stored in 'maps'
atlas_filename = atlas['maps']
# Loading atlas data stored in 'labels'
labels = atlas['labels']
# Load the functional datasets
data = datasets.fetch_development_fmri(n_subjects=1)
print('First subject resting-state nifti image (4D) is located at: %s' %
data.func[0])
Dataset created in /home/alexis/nilearn_data/development_fmri
Dataset created in /home/alexis/nilearn_data/development_fmri/development_fmri
Downloading data from https://osf.io/yr3av/download ...
...done. (1 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3df4712b400183b7092/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3e04712b400193b5bdf/ ...
...done. (2 seconds, 0 min)
First subject resting-state nifti image (4D) is located at: /home/alexis/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
Extract the time series#
from nilearn.maskers import NiftiMapsMasker
masker = NiftiMapsMasker(maps_img=atlas_filename, standardize=True,
memory='nilearn_cache', verbose=5)
masker.fit(data.func[0])
time_series = masker.transform(data.func[0],
confounds=data.confounds)
[NiftiMapsMasker.fit] loading regions from /home/alexis/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii
Resampling maps
/home/alexis/miniconda3/envs/nilearn/lib/python3.10/site-packages/nilearn/_utils/cache_mixin.py:304: UserWarning:
memory_level is currently set to 0 but a Memory object has been provided. Setting memory_level to 1.
________________________________________________________________________________
[Memory] Calling nilearn.image.resampling.resample_img...
resample_img(<nibabel.nifti1.Nifti1Image object at 0x7ff8fb20f0d0>, interpolation='continuous', target_shape=(50, 59, 50), target_affine=array([[ 4., 0., 0., -96.],
[ 0., 4., 0., -132.],
[ 0., 0., 4., -78.],
[ 0., 0., 0., 1.]]))
_____________________________________________________resample_img - 1.4s, 0.0min
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker._filter_and_extract...
_filter_and_extract('/home/alexis/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7ff8fb20f5e0>, { 'allow_overlap': True,
'detrend': False,
'dtype': None,
'high_pass': None,
'high_variance_confounds': False,
'low_pass': None,
'maps_img': '/home/alexis/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': True,
'standardize_confounds': True,
't_r': None,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/alexis/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=5)
[NiftiMapsMasker.transform_single_imgs] Loading data from /home/alexis/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
[NiftiMapsMasker.transform_single_imgs] Extracting region signals
[NiftiMapsMasker.transform_single_imgs] Cleaning extracted signals
_______________________________________________filter_and_extract - 1.7s, 0.0min
We can generate an HTML report and visualize the components of the
NiftiMapsMasker.
You can pass the indices of the spatial maps you want to include in the
report in the order you want them to appear.
Here, we only include maps 2, 6, 7, 16, and 21 in the report:
report = masker.generate_report(displayed_maps=[2, 6, 7, 16, 21])
report
/home/alexis/miniconda3/envs/nilearn/lib/python3.10/site-packages/numpy/ma/core.py:2826: UserWarning:
Warning: converting a masked element to nan.
time_series is now a 2D matrix, of shape (number of time points x number of regions)
print(time_series.shape)
(168, 39)
Build and display a correlation matrix#
from nilearn.connectome import ConnectivityMeasure
correlation_measure = ConnectivityMeasure(kind='correlation')
correlation_matrix = correlation_measure.fit_transform([time_series])[0]
# Display the correlation matrix
import numpy as np
from nilearn import plotting
# Mask out the major diagonal
np.fill_diagonal(correlation_matrix, 0)
plotting.plot_matrix(correlation_matrix, labels=labels, colorbar=True,
vmax=0.8, vmin=-0.8)
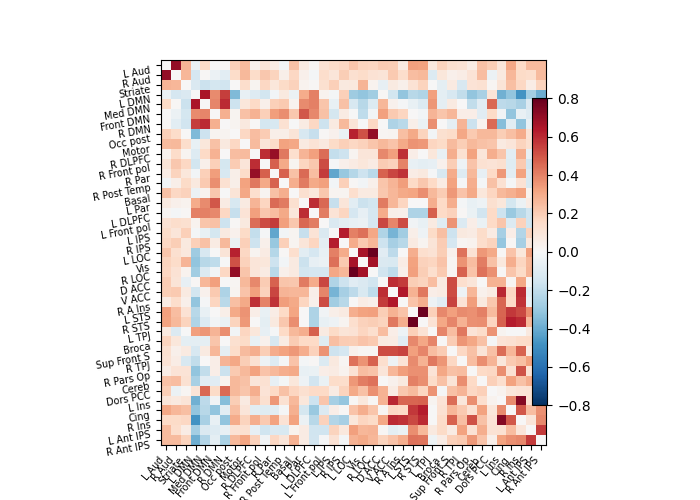
<matplotlib.image.AxesImage object at 0x7ff8ed826da0>
And now display the corresponding graph#
from nilearn import plotting
coords = atlas.region_coords
# We threshold to keep only the 20% of edges with the highest value
# because the graph is very dense
plotting.plot_connectome(correlation_matrix, coords,
edge_threshold="80%", colorbar=True)
plotting.show()
3D visualization in a web browser#
An alternative to nilearn.plotting.plot_connectome is to use
nilearn.plotting.view_connectome that gives more interactive
visualizations in a web browser. See 3D Plots of connectomes
for more details.
view = plotting.view_connectome(correlation_matrix, coords, edge_threshold='80%')
# In a Jupyter notebook, if ``view`` is the output of a cell, it will
# be displayed below the cell
view
# uncomment this to open the plot in a web browser:
# view.open_in_browser()
Total running time of the script: ( 0 minutes 16.549 seconds)
Estimated memory usage: 1066 MB