Note
Click here to download the full example code or to run this example in your browser via Binder
Breaking an atlas of labels in separated regions#
This example shows how to use
nilearn.regions.connected_label_regions
to assign each spatially-separated region of the atlas a unique label.
Indeed, often in a given atlas of labels, the same label (number) may be used in different connected regions, for instance a region in each hemisphere. If we want to operate on regions and not networks (for instance in signal extraction), it is useful to assign a different label to each region. We end up with a new atlas that has more labels, but each one points to a single region.
We use the Yeo atlas as an example for labeling regions,
nilearn.datasets.fetch_atlas_yeo_2011
The original Yeo atlas#
# First we fetch the Yeo atlas
from nilearn import datasets
atlas_yeo_2011 = datasets.fetch_atlas_yeo_2011()
atlas_yeo = atlas_yeo_2011.thick_7
# Let's now plot it
from nilearn import plotting
plotting.plot_roi(atlas_yeo, title='Original Yeo atlas',
cut_coords=(8, -4, 9), colorbar=True, cmap='Paired')
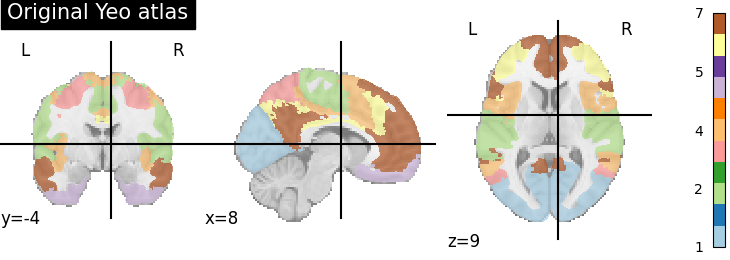
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7ff8eaef4190>
The original Yeo atlas has 7 labels, that is indicated in the colorbar. The colorbar also shows the correspondence between the color and the label
Note that these 7 labels correspond actually to networks that comprise several regions. We are going to split them up.
Relabeling the atlas into separated regions#
Now we use the connected_label_regions to break apart the networks of the Yeo atlas into separated regions
from nilearn.regions import connected_label_regions
region_labels = connected_label_regions(atlas_yeo)
/home/alexis/miniconda3/envs/nilearn/lib/python3.10/site-packages/nilearn/image/image.py:756: FutureWarning:
Image data has type int64, which may cause incompatibilities with other tools. This will error in NiBabel 5.0. This warning can be silenced by passing the dtype argument to Nifti1Image().
Plotting the new regions
plotting.plot_roi(region_labels, title='Relabeled Yeo atlas',
cut_coords=(8, -4, 9), colorbar=True, cmap='Paired')
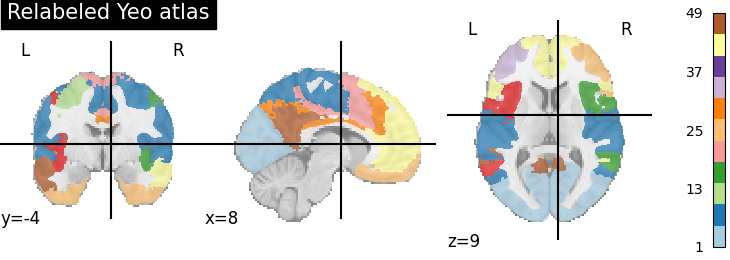
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7ff8eab2e860>
Note that the same cluster in original and labeled atlas could have different color, so, you cannot directly compare colors.
However, you can see that the regions in the left and right hemispheres now have different colors. For some regions it is difficult to tell apart visually, as the colors are too close on the colormap (eg in the blue: regions labeled around 3).
Also, we can see that there are many more labels: the colorbar goes up to 49. The 7 networks of the Yeo atlas are now broken up into 49 ROIs.
You can save the new atlas to a nifti file using to_filename method.
region_labels.to_filename('relabeled_yeo_atlas.nii.gz')
# The images are saved to the current folder. It is possible to specify the
# folder for saving the results, i.e.
# import os
# region_labels.to_filename(os.path.join(folder_path,
# 'relabeled_yeo_atlas.nii.gz'))
Different connectivity modes#
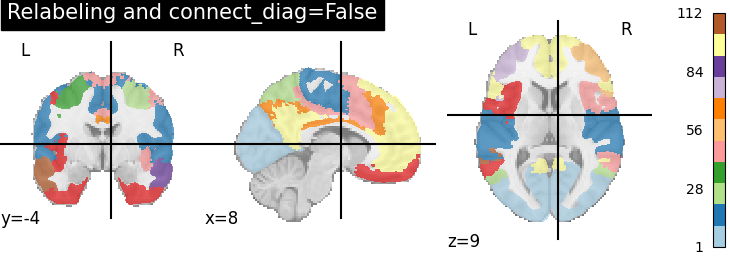
Using the parameter connect_diag=False we separate in addition two regions that are connected only along the diagonal.
region_labels_not_diag = connected_label_regions(atlas_yeo,
connect_diag=False)
plotting.plot_roi(region_labels_not_diag,
title='Relabeling and connect_diag=False',
cut_coords=(8, -4, 9), colorbar=True, cmap='Paired')
/home/alexis/miniconda3/envs/nilearn/lib/python3.10/site-packages/nilearn/image/image.py:756: FutureWarning:
Image data has type int64, which may cause incompatibilities with other tools. This will error in NiBabel 5.0. This warning can be silenced by passing the dtype argument to Nifti1Image().
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7ff8d645be80>
A consequence of using connect_diag=False is that we can get a lot of small regions, around 110 judging from the colorbar.
Hence we suggest use connect_diag=True
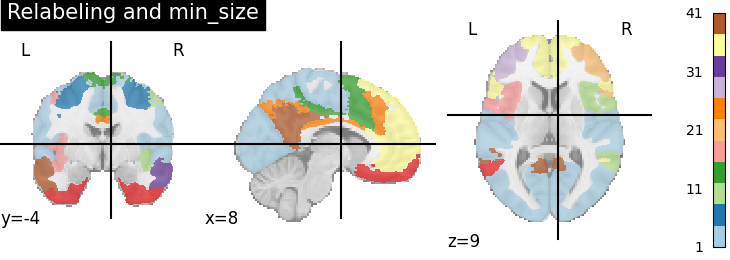
Parameter min_size#
In the above, we get around 110 regions, but many of these are very small. We can remove them with the min_size parameter, keeping only the regions larger than 100mm^3.
region_labels_min_size = connected_label_regions(atlas_yeo, min_size=100,
connect_diag=False)
plotting.plot_roi(region_labels_min_size, title='Relabeling and min_size',
cut_coords=(8, -4, 9), colorbar=True, cmap='Paired')
plotting.show()
/home/alexis/miniconda3/envs/nilearn/lib/python3.10/site-packages/nilearn/image/image.py:756: FutureWarning:
Image data has type int64, which may cause incompatibilities with other tools. This will error in NiBabel 5.0. This warning can be silenced by passing the dtype argument to Nifti1Image().
Total running time of the script: ( 1 minutes 25.911 seconds)
Estimated memory usage: 721 MB