Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.datasets.fetch_atlas_smith_2009#
- nilearn.datasets.fetch_atlas_smith_2009(data_dir=None, mirror='origin', url=None, resume=True, verbose=1)[source]#
Download and load the Smith ICA and BrainMap Probabilistic atlas (2009).
- Parameters
- data_dir
pathlib.Pathorstr, optional Path where data should be downloaded. By default, files are downloaded in home directory.
- mirror
str, optional By default, the dataset is downloaded from the original website of the atlas. Specifying “nitrc” will force download from a mirror, with potentially higher bandwidth. Default=’origin’.
- url
str, optional URL of file to download. Override download URL. Used for test only (or if you setup a mirror of the data). Default=None.
- resume
bool, optional Whether to resume download of a partly-downloaded file. Default=True.
- verbose
int, optional Verbosity level (0 means no message). Default=1.
- data_dir
- Returns
- data
sklearn.utils.Bunch Dictionary-like object, contains:
‘rsn20’:
str, path to nifti file containing the 20-dimensional ICA, resting-fMRI components. The shape of the image is(91, 109, 91, 20).‘rsn10’:
str, path to nifti file containing the 10 well-matched maps from the 20 maps obtained as for ‘rsn20’, as shown in 1. The shape of the image is(91, 109, 91, 10).‘bm20’:
str, path to nifti file containing the 20-dimensional ICA, BrainMap components. The shape of the image is(91, 109, 91, 20).‘bm10’:
str, path to nifti file containing the 10 well-matched maps from the 20 maps obtained as for ‘bm20’, as shown in 1. The shape of the image is(91, 109, 91, 10).‘rsn70’:
str, path to nifti file containing the 70-dimensional ICA, resting-fMRI components. The shape of the image is(91, 109, 91, 70).‘bm70’:
str, path to nifti file containing the 70-dimensional ICA, BrainMap components. The shape of the image is(91, 109, 91, 70).‘description’:
str, description of the atlas.
- data
Notes
For more information about this dataset’s structure: http://www.fmrib.ox.ac.uk/datasets/brainmap+rsns/
References
- 1(1,2,3)
Stephen M. Smith, Peter T. Fox, Karla L. Miller, David C. Glahn, P. Mickle Fox, Clare E. Mackay, Nicola Filippini, Kate E. Watkins, Roberto Toro, Angela R. Laird, and Christian F. Beckmann. Correspondence of the brain\textquoteright s functional architecture during activation and rest. Proceedings of the National Academy of Sciences, 106(31):13040–13045, 2009. URL: https://www.pnas.org/content/106/31/13040, arXiv:https://www.pnas.org/content/106/31/13040.full.pdf, doi:10.1073/pnas.0905267106.
- 2
Angela R. Laird, P. Mickle Fox, Simon B. Eickhoff, Jessica A. Turner, Kimberly L. Ray, D. Reese McKay, David C. Glahn, Christian F. Beckmann, Stephen M. Smith, and Peter T. Fox. Behavioral Interpretations of Intrinsic Connectivity Networks. Journal of Cognitive Neuroscience, 23(12):4022–4037, 12 2011. URL: https://doi.org/10.1162/jocn\_a\_00077, arXiv:https://direct.mit.edu/jocn/article-pdf/23/12/4022/1777164/jocn\_a\_00077.pdf, doi:10.1162/jocn_a_00077.
Examples using nilearn.datasets.fetch_atlas_smith_2009#
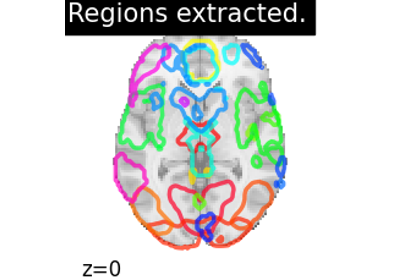
Regions Extraction of Default Mode Networks using Smith Atlas