Note
Click here to download the full example code or to run this example in your browser via Binder
Visualizing global patterns with a carpet plot#
A common quality control step for functional MRI data is to visualize the data over time in a carpet plot (also known as a Power plot or a grayplot).
The nilearn.plotting.plot_carpet function generates a carpet plot
from a 4D functional image.
Fetching data from ADHD dataset#
from nilearn import datasets
adhd_dataset = datasets.fetch_adhd(n_subjects=1)
# plot_carpet can infer TR from the image header, but preprocessing can often
# overwrite that particular header field, so we will be explicit.
t_r = 2.
# Print basic information on the dataset
print('First subject functional nifti image (4D) is at: %s' %
adhd_dataset.func[0]) # 4D data
/home/alexis/miniconda3/envs/nilearn/lib/python3.10/site-packages/nilearn/datasets/func.py:250: VisibleDeprecationWarning: Reading unicode strings without specifying the encoding argument is deprecated. Set the encoding, use None for the system default.
phenotypic = np.genfromtxt(phenotypic, names=True, delimiter=',',
First subject functional nifti image (4D) is at: /home/alexis/nilearn_data/adhd/data/0010042/0010042_rest_tshift_RPI_voreg_mni.nii.gz
Deriving a mask#
from nilearn import masking
# Build an EPI-based mask because we have no anatomical data
mask_img = masking.compute_epi_mask(adhd_dataset.func[0])
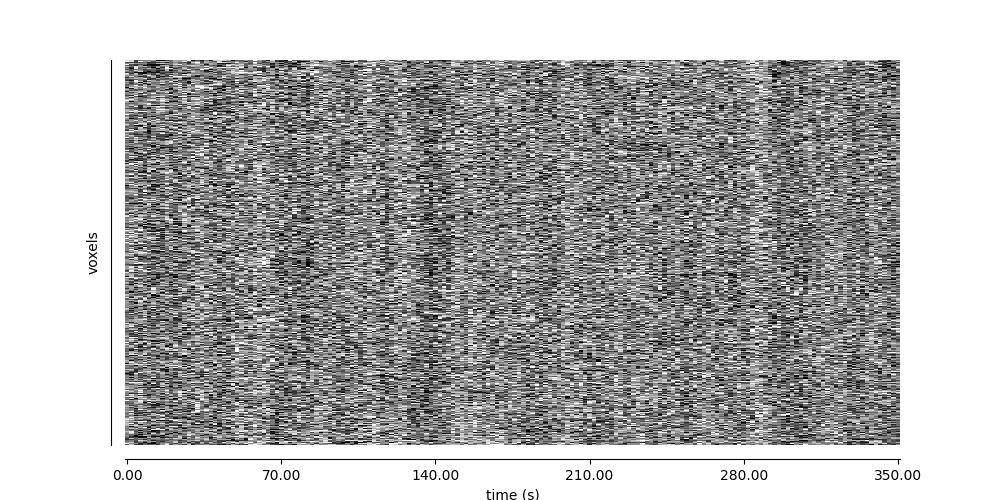
Visualizing global patterns over time#
import matplotlib.pyplot as plt
from nilearn.plotting import plot_carpet
display = plot_carpet(adhd_dataset.func[0], mask_img, t_r=t_r)
display.show()
Deriving a label-based mask#
Create a gray matter/white matter/cerebrospinal fluid mask from ICBM152 tissue probability maps.
import numpy as np
from nilearn import image
atlas = datasets.fetch_icbm152_2009()
atlas_img = image.concat_imgs((atlas["gm"], atlas["wm"], atlas["csf"]))
map_labels = {"Gray Matter": 1, "White Matter": 2, "Cerebrospinal Fluid": 3}
atlas_data = atlas_img.get_fdata()
discrete_version = np.argmax(atlas_data, axis=3) + 1
discrete_version[np.max(atlas_data, axis=3) == 0] = 0
discrete_atlas_img = image.new_img_like(atlas_img, discrete_version)
/home/alexis/miniconda3/envs/nilearn/lib/python3.10/site-packages/nilearn/image/image.py:756: FutureWarning: Image data has type int64, which may cause incompatibilities with other tools. This will error in NiBabel 5.0. This warning can be silenced by passing the dtype argument to Nifti1Image().
return klass(data, affine, header=header)
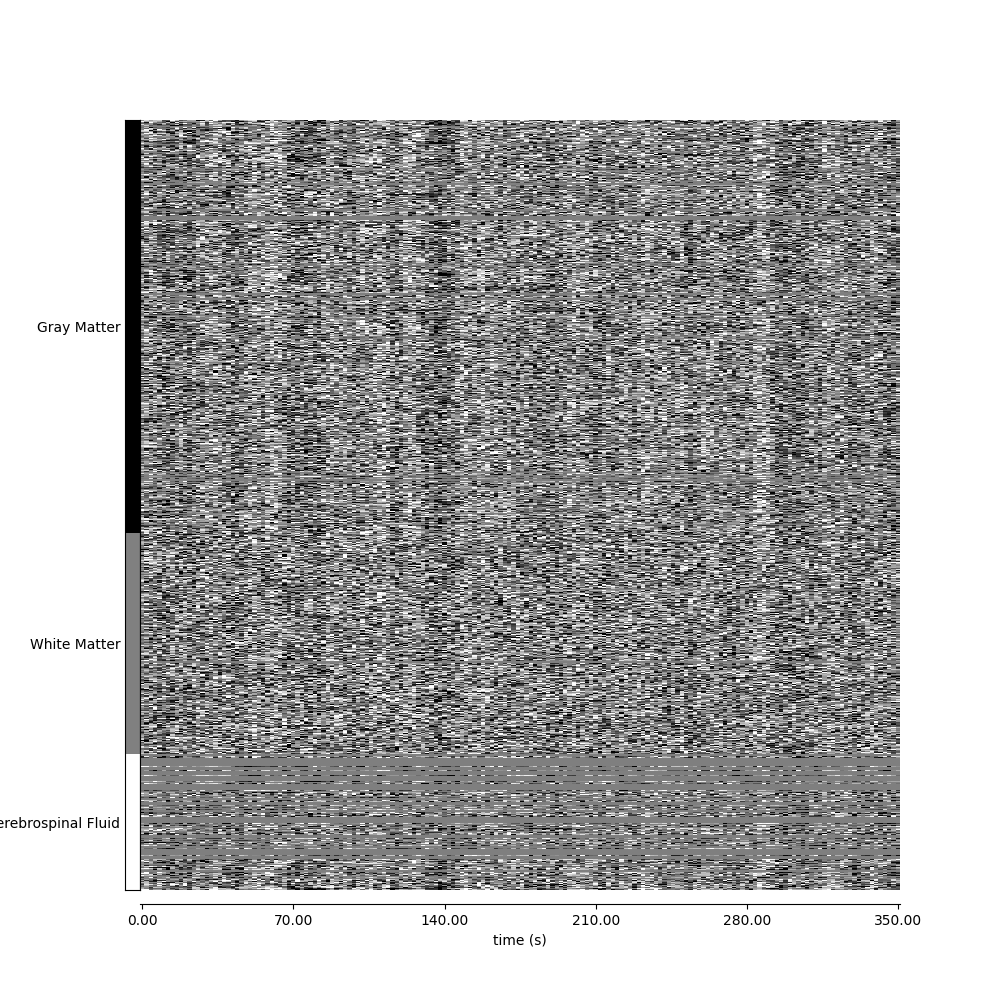
Visualizing global patterns, separated by tissue type#
import matplotlib.pyplot as plt
from nilearn.plotting import plot_carpet
fig, ax = plt.subplots(figsize=(10, 10))
display = plot_carpet(
adhd_dataset.func[0],
discrete_atlas_img,
t_r=t_r,
mask_labels=map_labels,
axes=ax,
cmap="gray",
)
fig.show()
/home/alexis/miniconda3/envs/nilearn/lib/python3.10/site-packages/nilearn/image/resampling.py:531: UserWarning: Casting data from int32 to float32
warnings.warn("Casting data from %s to %s" % (data.dtype.name, aux))
Coercing atlas_values to <class 'int'>
Total running time of the script: ( 0 minutes 8.468 seconds)
Estimated memory usage: 1042 MB